Genomic sequencing of mutated bacteria becomes possible in 2 days
성수 최 2009-09-29 View. 16,799Genomic sequencing of mutated bacteria becomes possible in 2 days
Scientists identify mechanism for emergence of mutated cholera bacterium Working through an international study, Korean scientists have identified the mechanism through which pandemic cholera bacterium evolves - a world's first.
The Ministry of Education, Science and Technology (Minister Ahn Byung-man) said on August 31 that the research teams led by Prof. Chun Jong-sik at Seoul National University and Dr. Kim Dong-wook at the International Vaccine Institute (IVI) analyzed 23 cholera biotypes, becoming the first in the world to identify the cause for the emergence of a mutated bacterium and how pathogenic bacteria evolve.
The study was conducted jointly by researchers in three countries, namely, the U.S. (University of Maryland), Korea and India. The study was published in the online edition of the Proceedings of National Academy of Sciences.

(Photo) The two-chromosome cholera bacterium genome, and the location and size of the 70 genetic groups moving in between genes, which were discovered in this study. Variants consist of different combinations of those genetic groups. According to the study, variants are believed to be generated due to bacteriophage, the virus that infects bacteria.
The study is the largest on genomic analysis of microorganisms ever publicized in the world. The University of Maryland and the Department of Energy Joint Genome Institute at the Los Alamos National Laboratory in the U.S. conducted gene sequencing of Vibrio cholerae, while the SNU and IVI teams took charge of bioinformatics analysis. Prof. Chun conducted the study with financial support from the "National Research Laboratory Program", which was funded by MEST and the Korea Research Foundation (President Park Chan-mo).
'Bacteriophage'spawned evolution of cholera bacteria.
Cholera is a diarrheal disease caused by the Vibrio cholerae bacteria. It is a deadly infectious disease which, if left untreated, can kill a patient in a matter of 18 hours.?? According to the World Health Organization, cholera infected 170,000 people in 53 countries and killed 4,031 people in 2007. If underreported cholera deaths are taken into consideration, the disease kills an estimated 120,000 people a year.
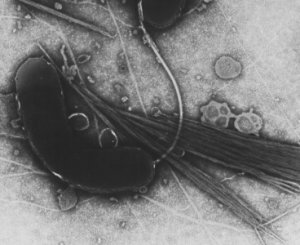
(Photo) Electron microscopic picture of Vibrio cholerae, the bacterium that causes cholera, an infectious diarrheal disease fatal to humans.
In Korea, there is a growing chance of cholera outbreaks due to global warming and a rise in seawater temperature; thus, cholera is strictly managed as a Grade 1 infectious disease by health authorities. Cholera is also categorized as a Category B bioterrorism pathogen in the United States.
The joint team of international researchers conducted genomic sequencing of 23 V. cholerae strains that had been collected from around the world since 1910, and analyzed them using the latest bioinformatics techniques. The researchers found that the transition from the sixth to seventh pandemic involved a transformation of the pathogen into a wholly new strain, and that different strains that emerged or are emerging during the current seventh pandemic are clones that recently evolved from the same ancestor.
The researchers concluded that mutated bacteria emerging every several years are spawned by bacteriophage, the virus that infects bacterium. They found that many of the genes that play key roles in triggering diseases, including cholera toxic, a main culprit of diarrhea, are transferred between bacteria due to this virus. 6,000 genes from the 23 V. cholerae stains discovered.
Unlike humans, bacteria can transmit and receive genes between themselves. The researchers identified about 70 new genetic groups that move between bacteria. They found that as bacteria are created in the body in various combinations of those genetic groups, new mutated bacteria strains emerge. The identification of the evolutionary mechanism of pathogenic bacteria has paved the way to promptly cope with newly emerging strains of bacteria, and to efficiently develop vaccines and therapeutics.
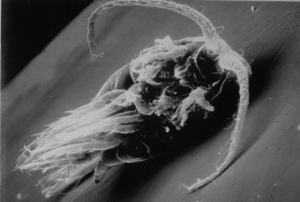
(Photo) Copepod, a type of animal plankton, on which cholera bacterium often resides. According to the study, cholera bacteria concentrate on the surface of this plankton, where they exchange genes between themselves.
In the study, the researchers discovered as many as 6,000 genes from the 23 V. cholerae stains. The researchers believe that the entire genetic pool of cholera bacteria that exist in nature contains hundreds of thousands of genes. Accordingly, they predict that a more virulent and infectious mutated bacterium can emerge any time.
The study, the largest in scale among studies on microorganism genomes ever publicized, also provides an important clue to the mechanism of how new strains emerge in other pathogenic bacteria, including anthrax, shigella, typhoid, helicobacter pylori, and streptococcus pneumoniae. Moreover, by using the database developed through the study, scientists can now diagnose a mutated cholera strain that emerges anywhere in the world, including Korea, in a matter of 48 hours through genomic analysis.
Prof. Chun, who led the study, explained the significance of the findings, saying that "By discovering the evolutionary mechanism of the cholera bacterium - the cause of a disease that was difficult to efficiently control due to its frequent mutations -? the study has laid a scientific foundation and serves as a milestone that is essential to accurate diagnosis and vaccine development." He added, "By taking the lead in the development of a genomic database for major infectious pathogens, Korea has established the groundwork to effectively cope with a national crisis amid a growing possibility for cholera outbreaks in Korea due to global warming." Genome and Lateral gene transfer
A genome is a constellation of genes, and is a blueprint of a given organism. The human genome consists of 3 billion bases, while the cholera bacterium genome comprises about 4 million. Humans have about 25,000 genes, while the cholera bacterium contains 4,000.
Humans have identical genes, while different cholera strains have genes that can be different by a factor of up to 10 percent. Bacteria have the capacity to lose genes and acquire new ones from other bacterium, a phenomenon called 'Lateral gene transfer'. Cholera bacterium, Vibrio cholerae
Cholera is a fatal diarrheal disease that is caused by Vibrio cholerae, a gram negative bacterium. The disease is prevalent in Asia and Africa, primarily near the tropical region. Recently, a major outbreak in southern Africa killed thousands of people.
Cholera causes illness only in men, while swine cholera, which afflicts pigs, is a disease that is caused by a virus, which is a totally different microorganism.
Cholera is highly infectious in humans, but Vibrio cholerae resides primarily in seawater. Notably, the bacterium are found at the mouth of rivers where freshwater meets the ocean and where the salinity level of seawater declines. It is known to? attach itself to animal plankton, rather than residing in the water itself.
According to a study published in the journal 'Nature' in 2000 by Gregory M. Luiz of the Smithsonian Environmental Institute, cholera was found in 93 percent of the ballast water (water contained in vessels to keep the balance of the vesselas buoyancy) in freight ships navigating through the Chesapeake Bay in the United States, As the cholera bacterium can spread via freight ships navigating across different continents, the International Maritime Organization enacted an international treaty on ballast water management, which calls for the level of cholera contamination in the water to be less than one bug per 100 ml of water. (http://www.imo.org/Conventions/mainframe.asp?topic_id=867)
?
Kim Chung-han
chkim at kofac.or.kr
[September 15, 2009]
- - - - - - -
Source - ScienceTimes

 Delete Article!
Delete Article!