KISTI achieves Parallelization of world-class Genome Wide Association Study(GWAS)
이정훈 2022-04-07 View. 109,704KISTI achieves Parallelization of world-class Genome Wide Association Study(GWAS)
- Optimization of Genome Wide Association Study's statistical error correction through parallel computing -
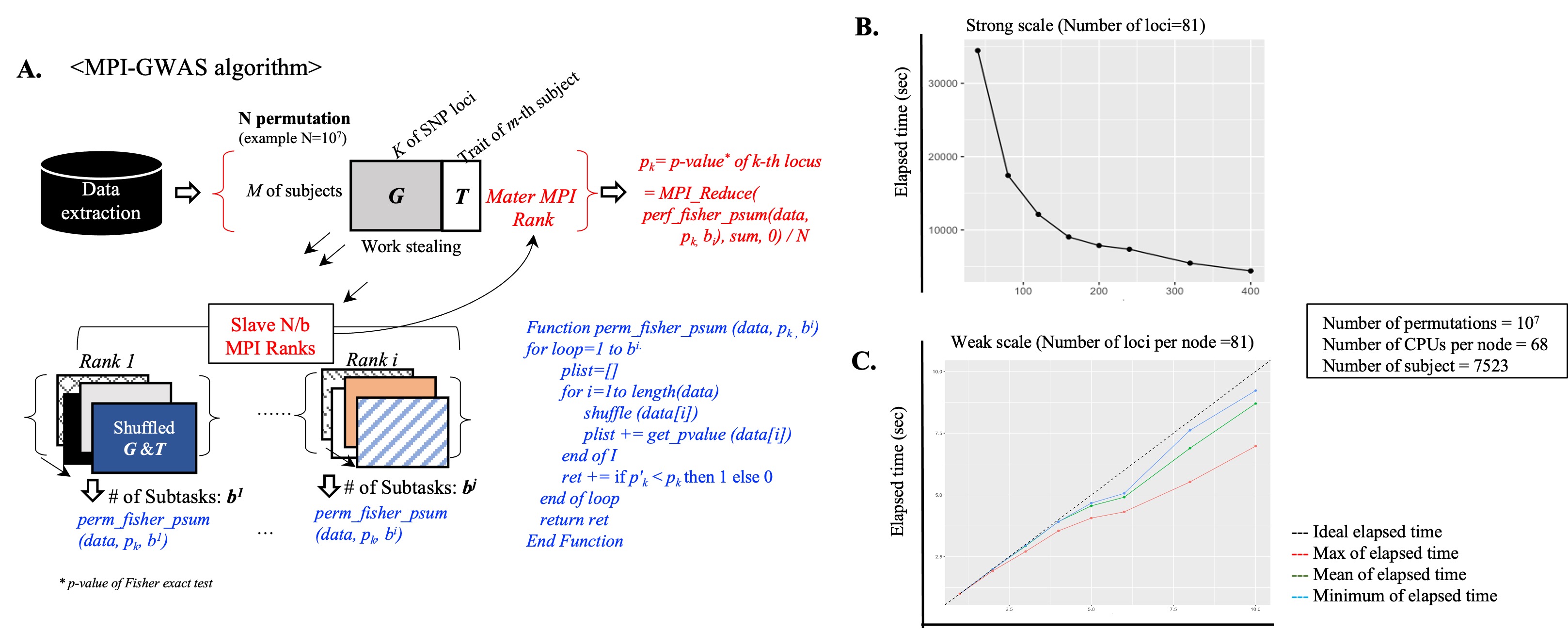
A. Summary of the MPI-GWAS algorithm: Simultaneous/parallel analysis through MPI-Rank for quick calculation of N-Permutations
B-C. Performance evaluation results:
(B) Decrease in overall elapsed time by increasing parallel processing nodes
(C) Expandability of elapsed time as simultaneous increasing with parallel processing nodes and data
KISTI announced that it has developed a large-scale supercomputing software to correct statistical errors* in Genome Wide Association Study(GWAS).
* It refers to a phenomenon in which the statistical significance calculation result through the large-scale GWAS is reported as false-positive. Representative cases of false-positive in the field of GWAS are that genetic mutations with low posibility of a genetic disease are reported to be highly related to the disease.
Using the SW developed at Nurion, KISTI's 5th National Supercomputer, genetic mutations related to diabetes and high blood pressure from repoted 84,295 genetic mutations in 7,523 Korean cohorts and 4,242 British cohorts, were derived through GWAS calculations, and corrected those statistical errors by performing up to 7 billion random permutations.
The supercomputing simulation SW can accelerate calculations by more than 300% compared to existing statistical programs through using up to 2,500 nodes* of Nurion.
* It is capable of performing about 7.5 petaflops (1 petaflops = 1,000 trillion instructions per second) and accounts for about 25% of the performance of Nurion.
The results of the GWAS analysis aim to select phenotype variation(disease or fruit weight, etc.), and the discovery of significant disease-related genetic mutaions is considered an important indicator of personalized health care and new varieties improvement in agriculture. Therefore, statistical error correction of GWAS results is essential.
The statistical error correction of GWAS remained a challenge in the field because of its vast calculation. KISTI confirmed that it is possible to correct the existing errors by large-scale supercomputing-based petaflops-class calculating through parallel computing technology. Using Nurion, KISTI performed on GWAS with the world-class resources(7.5 petaflops).
Dr. Kwon Oh-kyung and Dr. Paik Hyo-jung said, "This GWAS parallelization SW has been released its source code*, so that various genetic researchers can freely use it. We are expecting a research efficiency using supercomputer in the genetic field."
Its research was published on March 31st in Genomics & Informatics**.
* https://github.com/hypaik/proj_MPIGWAS
** Paik et al, MPI-GWAS: a supercomputing-aid permutation approach for genome-wide association studies
Jeong Min-joong, director of Supercomputing Application Center said,
"KISTI supports optimal parallelization* technology and computational resources for supercomputer users who need large-scale calculation. We expect that the distributed supercomputing simulation software will provide the research efficiency in the bio and medical fields."
* Optimal parallelization is a technology that solves difficult problems by developing code that allows thousands of CPUs to perform simultaneously on supercomputer.
Attached files(1)

 Delete Article!
Delete Article!